Abstract
Substance use disorders are complex biopsychosocial disorders that have substantial negative neurocognitive impact in various patient populations. These diseases involve the compulsive use of licit or illicit substances despite adverse medicolegal consequences and appear to be secondary to long-lasting epigenetic and transcriptional adaptations in brain reward and non-reward circuits. The accumulated evidence supports the notion that repeated drug use causes changes in post-translational histone modifications and in DNA methylation/hydroxymethylation processes in several brain regions. This review provides an overview of epigenetic changes reported in models of cocaine, methamphetamine, and opioid use disorders. The accumulated data suggest that future therapeutic interventions should focus on the development of epigenetic drugs against addictive diseases.
1. Background and introduction
Diagnostic criteria that are necessary to meet a diagnosis of substance use disorder (SUD) include compulsion to use licit or illicit substances and continuous use in the presence of adverse medical, neurological and/or psychiatric complications (DSM5, 2013). SUDs that are associated with increased morbidity and mortality in susceptible individuals (Degenhardt et al., 2014) are thought to be secondary to maladaptive changes in several brain regions [(Volkow and Boyle, 2018) see also Fig. 1]. Importantly, negative cognitive impact of substance use suggests that therapeutic approaches against SUDs will only be successful if and only if they address the various clinical courses associated with various patient populations because the clinical outcomes of patients are not uniform (Soares and Pereira, 2019). It is to be noted, at the onset, that neuroadaptations and their clinical sequelae are influenced by genetic and environmental factors that also account for epigenetic and transcriptional responses to these drugs in the central nervous system (Fig. 1). Using similar reasoning, several groups of investigators have tried to mimic these disorders in animal models in attempts to develop better pharmacological approaches to treat patients with SUDs (Cadet, 2019). Some of these models include conditioned place preference (CPP) and drug self-administration (SA) (Cadet et al., 2019; Muller, 2018). Molecular studies using various animal models have now provided evidence for the participation multiple gene networks in the development and maintenance of drug taking behaviors (Cadet et al., 2015; Korostynski et al., 2013; Nestler, 2012).
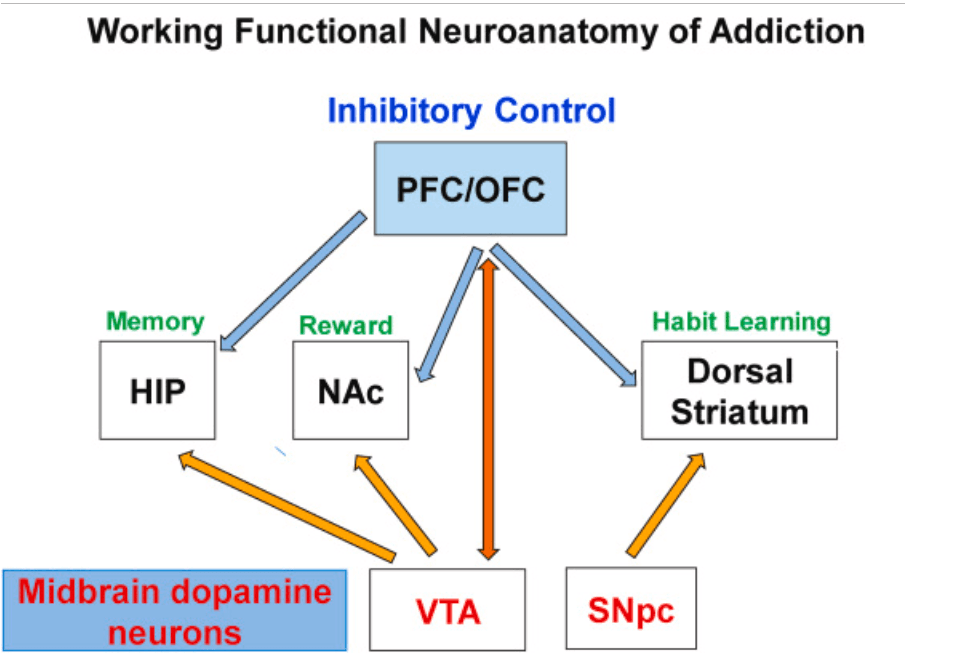
Fig. 1. This figure provides a working model of brain regions that are presumed to be involved in the development and maintenance of addictive states. Although the figure shows mainly dopaminergic projections that participate in reward circuits, there are non-reward projections that definitely participate in the maintenance of addiction because these diseased states involved more than rewarding effects of drugs. Future studies will need to emphasize more complex interactions of reward and non-reward pathways in the clinical course of addiction in individual patients and their responses to therapeutic interventions.
2. Epigenetic regulation of gene expression
Changes in gene expression are regulated by epigenetic changes that include post-translational histone modifications, DNA methylation, and microRNAs (Jackson and Standart, 2007; Mazzio and Soliman, 2012; Murr, 2010). Modifications of N-tails of histones that possess lysine residues include acetylation/deacetylation processes catalyzed reversibly by histone acetyltransferases (HATs) and histone deacetylases (HDACs) (Falkenberg and Johnstone, 2014; Voss and Thomas, 2018). There are three classes of nuclear HATs that have histone acetyltransferase domains and bind acetyl-coenzyme A (Voss and Thomas, 2018). They comprise the GNAT (GCN5-related acetyltransferase), CBP/p300, and the MYST (MOZ, Ybf2/Sas 3, Sas 2, and Tip 60) families of enzymes. HDACs are divided into four classes based on sequence similarities (Mottet and Castronovo, 2008; Yoshida et al., 2017). Those class I (HDAC1, HDAC2, HDAC3, and HDAC8), class IIA (HDAC4, HDAC5, HDAC7, HDAC9), class IIB (HDAC6, HDAC10), class III (Sirtuins 1-7) and Class IV (HDAC11) HDACs. Class I, II and IV HDACs are the classical HDACs that are Zn2+-dependent enzymes whereas the sirtuins require NAD+-dependent.
Histone methylation, catalyzed by lysine methyltransferases (KMTs), is also an important player in the regulation of gene expression (Black et al., 2012; Hyun et al., 2017). These enzymes include KMT1A (SUV39H1), KMT1B (SUV39H2), KMT1C (G9a), KMT2A (MLL1), KMT2B (MLL2), KMT3A (SET2), KMT3B (NSD1), among others. In addition to the KMTs, protein arginine N-methyltransferases (PRMTs) can catalyze the addition of methyl groups to arginine residues in histones (Cha and Jho, 2012; Wolf, 2009).
Gene expression in the brain is also regulated by DNA methylation (Bayraktar and Kreutz, 2018). DNA contains four bases that are adenine (A), guanine (G), cytosine (C) and thymine (T). Cytosine can be altered at the carbon-5 position by adding a methyl group to form 5-methylcytosine (Jones, 2012; Moore et al., 2013). Mammalian species have enzymes named DNA methyltransferases (DNMTs) (DNMT1, DNMT3A, DNMT3B) that catalyze these reactions (Lyko, 2018). Methylated CpG pairs are found throughout the genome and DNA methylation can participate in repression of transcription (Moore et al., 2013). Methylated DNA can also be oxidized to form hydroxymethylated DNA by ten-eleven translocation (TET) enzymes (Lio et al., 2020). In addition to DNA hydroxymethylation, these enzymes are also involved in the formation of 5-formylcytosine, and 5-carboxylcytosine. TET enzymes and DNA hydroxymethylation have been reported to participate in the regulation of plasticity-related genes, learning, and memory (Kaas et al., 2013; Kumar et al., 2015) that are important to the addiction process.
3. Models of drug administration and post-translational histone acetylation
Because cocaine can alter gene expression (Bannon et al., 2005; Nestler et al., 1993; Yano and Steiner, 2007), several groups of investigators have sought to determine how the drug might impact histone modifications and HAT expression in various brain regions (Levine et al., 2005). were the first to show that cocaine increased the binding of CBP, a histone acetyl-transferase (Bannister and Kouzarides, 1996) that is involved in the regulation of many cellular events (Goodman and Smolik, 2000, Lipinski et al., 2019), at the fosB promoter accompanied by increased fosB mRNA expression. Acute and chronic non-contingent cocaine administration by experimenters caused increased acetylation of histone H3 at lysine 14 (H3K14Ac) and of histone H2B at lysine 12 (H2BK12Ac) in the nucleus accumbens (NAc) of mice (Malvaez et al., 2011), changes that were attenuated in mice with CBP deletion in the NAc. Bilateral deletion of CBP in the NAc also attenuated cocaine conditioned place preference (CPP). Interestingly, however, Madsen et al. (2012) showed that deletion of CBP in the mouse striatum produced increased sensitivity to cocaine and amphetamine.
In addition to experiments using cocaine administered by experimenters, some papers have also used studies in which rodents self-administered cocaine (Freeman et al., 2008; Monsey et al., 2019; Sadakierska-Chudy et al., 2017a; Sadri-Vakili et al., 2010). Freeman et al. (2008) used discrete-trials cocaine self-administration (SA) after rats had established a stable daily pattern of cocaine intake for 5 days (40 infusions within 6 h). During those session, rats could access cocaine during 10-min discrete trials. A trial ended when a cocaine injection (1.5 mg/kg/injection) was collected or 10 min had elapsed. Rats received four discrete trials per hour (i.e., DT4), 24 h per day for 10 days. The authors found that forced abstinence from cocaine self-administration (SA) for one and 10 days was associated with decreased abundance of H3 acetylated at K9 and K14 bound at the EGR1 promoter and decreased EGR1 mRNA levels but increased H3K9/14ac at the promoter gene of Npy and increased NPY expression in the rat mPFC after one day of abstinence. Interestingly, Sadri-Vakili et al. (2010) reported that cocaine SA followed by 7 days of forced abstinence also increased abundance of acetylated H3K9/14Ac at the Bdnf IV promoter and increased mRNA expression in rat medial prefrontal cortex (mPFC). Another group of investigators trained rats to self-administer cocaine using 2-h daily sessions 6 days/week for a minimum of 12 days (Sadakierska-Chudy et al., 2017a). At the end of the experiments, microarray analyses were used to measure gene expression in the rat prefrontal cortex (PFC). They found that mRNA levels of several epigenetic enzymes were significantly impacted in the PFC of rats that had self-administered cocaine. They also found significant increases in acetylated histone H3K9 and H4K8 but not H3K27 or H4K5 (Sadakierska-Chudy et al., 2017a). The role of HATs in cocaine-associated behaviors was also tested in cocaine SA experiments by Monsey et al. (2019). In those experiments, rats underwent 12 days of cocaine self-administration (SA) training using 1-h sessions. Monsey et al. (2019) reported that injections of garcinol, an inhibitor of KATs of p300/CBP and PCAF families (Arif et al., 2009; Balasubramanyam et al., 2004), into the lateral nucleus of the amygdala (LA) was able to block the reconsolidation of a cocaine-cue memory. They also reported that systemic injections of garcinol caused decreased abundance of acetylated histone H3K27 in the LA. Acetylation of histone H3K27 is known to be mediated, in part, by CBP/p300 (Jin et al., 2011). Together, these data suggest that the involvement of identifiable epigenetic events in cocaine-induced behaviors may be specific to certain brain regions.
Injection of another psychostimulant, methamphetamine, also causes changes in histone acetylation (Martin et al., 2012). Methamphetamine injection caused time-dependent increases in acetylated H4K5 (H4K5Ac) and H4K8 (H4K8Ac) levels that were associated with increased expression of the histone acetyltransferase, ATF2. A follow-up study found that acute and chronic METH injections could cause differential changes in striatal gene expression that was associated with increased H4K5Ac binding near the transcriptional start sites (TSSs) of several genes using chromatin immunoprecipitation followed by genome wide sequencing (Cadet et al., 2013). There were also significant positive correlations between gene expression obtained by microarray analysis and H4K5Ac binding by ChIP-Seq in the striatum of control and METH-treated rats. Genes of interest that show increased mRNA levels and H4K5Ac binding include immediate early genes (IEGs) such as Arc, c-fos, Egr 2, Egr 4, JunB, Nr4a3, and Npas4 (Cadet et al., 2013). Other genes of interest are DUSP5, DUSP 14, Neurotensin, and Per 1 (Cadet et al., 2013). These results implicate this epigenetic marker in the molecular effects of methamphetamine in the brain. Those results also suggest that HAT1/KAT1 which is responsible for histone H4 acetylation (Nagarajan et al., 2013, Parthun, 2007) in the long-term effects of this drug since repeated injections of METH are also associated with increased H4K5 acetylation (Cadet et al., 2013). Moreover, Gonzalez et al. (2019) reported that a single injection of METH increased the abundance of acetylated histone H4 on Hdac1 and Hdac10 gene promoters. They also found decreased acetylated H4 on Hdac4 and Hdac5 genes. In contrast, they found that repeated injections of METH increased H4 acetylation on Hdac4 and Hdac5 genes but decreased abundance on Hdac1, Hdac2, and Hdac8 gene promoters. More studies are necessary to further test the role of HATs and histone acetylation in models that use methamphetamine self-administration.
There is evidence that class I HDACs are also differentially regulated in animal models of drug administration. Schroeder et al. (2008) showed that pretreatment of mice with the class I inhibitor, sodium butyrate, in conjunction with a dopamine D1 agonist caused increased TH and BDNF mRNA expression in the ventral midbrain, changes that were associated, unexpectedly, with H3 hypoacetylation at the promoters of these genes. Kennedy et al. (2013) showed that local knockdown of HDAC1, but not of HDAC2 and HDAC3, caused reduction of cocaine-induced locomotor sensitization. In addition, Host et al. (2011) reported that cocaine SA increased HDAC2 mRNA expression in the NAc, dorsal striatum and cingulate cortex. Importantly, reduction of HDAC2 expression by induction of cyclic GMP reduced cocaine SA (Deschatrettes et al., 2013). Another class I HDAC, HDAC3, appears to also participate in cocaine-induced behaviors. Specifically, systemic administration of RGFP966, a selective HDAC3 inhibitor, was able to facilitate extinction of cocaine CPP while increasing acetylation of histone H4 at lysine 8 in the infra-limbic cortex (Malvaez et al., 2013). RGFP increased H3K14 acetylation in the infra-limbic cortex, NAc and hippocampus (Malvaez et al., 2013). RGFP966 treatment decreased HDAC3 binding at the c-fos promoter (Malvaez et al., 2013). Methamphetamine, another illicit drug, also impacts the expression of class I HDACs. Martin et al. (2012) reported that a single methamphetamine injection decreased HDAC1 but increased HDAC2 protein levels in the rodent NAc. In addition, Jayanthi et al. (2014) showed that repeated injections of methamphetamine increased HDAC1 and HDAC2 expression in the dorsal striatum. These increases were associated with hypoacetylation of histone H4 at lysines 4, 8, 12 and 16 (Jayanthi et al., 2014).
Class II HDACs are also involved in the behavioral effects of drugs of abuse. For example, chronic bilateral injection of the HDAC inhibitor, Trichostatin (TSA), in the rat NAc shell caused higher SA rates in rats by influencing, in part, HDAC4 expression because HDAC4 overexpression attenuated the motivation to self-administer cocaine (Wang et al., 2010) Chronic non-contingent cocaine injections decreased HDAC5 expression in the NAc (Renthal et al., 2007) whereas cocaine SA decreased nuclear HDAC5 localization (Host et al., 2011). HDAC5 knockout in rodents increased cocaine CPP and that viral HDAC5 overexpression in the NAc decreased cocaine CPP (Renthal et al., 2007). Dietrich et al. (2012) also showed that repeated cocaine injections induced salt-inducible kinase (SIK1)-dependent HDAC5 phosphorylation in the nucleus and its subsequent shuttling to the cytoplasm. In addition to cocaine, methamphetamine can also impact the expression of these HDACs. Specifically, an acute methamphetamine injection decreased HDAC4 and HDAC7 mRNA levels in the NAc of WT mice (Torres et al., 2016). In contrast, Li et al. (2015) observed significant increases in the mRNA expression of HDAC4 and HDAC5 in the dorsal striatum of rats that had undergone 35 days of withdrawal from methamphetamine SA. Furthermore, striatal HDAC5 overexpression or knockdown respectively increased or decreased methamphetamine seeking after a long interval of withdrawal (Li et al., 2017).
The use of psychostimulants also impacts the expression of class III HDACs that are the SIRTs (Satoh et al., 2017). Renthal et al. (2009) performed chromatin immunoprecipitation (ChIP) to investigate the effects of repeated injections of cocaine in the NAc of mice and found that cocaine increased histone acetylation at Sirt1, and Sirt2 DNA sequences (Renthal et al., 2009). Ferguson et al. (2013) also reported that overexpression of Sirt1 in the NAc increased cocaine CPP whereas its depletion caused decreased CPP (Ferguson et al., 2013). Interestingly, although cocaine administration increased SIRT1 induction in the rodent nucleus accumbens, Ferguson et al. (2015) found that SIRT1 abundance was decreased at gene promoters. These cocaine-induced changes were also accompanied by increased expression of SIRT1 target genes. The authors found that increased SIRT1 expression was associated with deacetylation of FOXO3a and increased expression of known FOXO3a gene targets. Consistent with the results with cocaine, repeated methamphetamine injections also increased Sirt1 and Sirt2 mRNA and protein levels in the rat dorsal striatum (Jayanthi et al., 2014). Consistent with these observations, chronic METH was recently found to increase H4 acetylation binding at the Sirt2 promoter (Gonzalez et al., 2020). These findings thus implicate class III HDACs in the regulation of some genes that might be involved in mediating long-term neuroadaptations to exposure to psychostimulants.
The class IV HDAC, HDAC11, which was cloned and characterized by Gao et al. (2002), is highly expressed in the brain (Takase et al., 2013). Similar to the other HDACs, Host et al. (2011) have reported that cocaine self-administration was accompanied by increased HDAC11 expression. In contrast, acute METH injections caused decreases in HDAC11 mRNA levels in the NAc (Omonijo et al., 2014; Torres et al., 2016).
In addition to the effects of psychostimulants on histone acetylation/deacetylation, opioids have also been reported to cause histone post-translational modifications (Browne et al., 2020). Sheng et al. (2011) had demonstrated that heroin administration was associated with increased histone H3 phosphoacetylation that was essential to heroin CPP. These changes in histone acetylation observed in rodents are consistent with a report of increased acetylation of H3K27 in the post-mortem striatum of heroin addicts (Egervari et al., 2017). Importantly, HDAC inhibition was able to enhance morphine-associated memory formation (Wang et al., 2015), further suggesting a role of histone acetylation in opioid-mediated behaviors.
4. Models of drug administration and post-translational histone methylation
Cocaine-induced changes in gene expression appear to also depend on alterations in histone methylation although this histone mark has been less well investigated in models of addiction. Maze et al. (2010) reported that repeated injections of cocaine reduced global levels of H3K9 dimethylation in the NAc. These decreases were due to G9a repression in a deltaFoB-dependent fashion. They further showed that viral-mediated G9a down-regulation enhanced preference for cocaine. In addition, Jordi et al. (2013) reported that a single cocaine injection induced significant changes in histone methylation at 15 min after the drug administration. Histone H3K9 trimethylation was increased in D1-containing neurons whereas all three methylated forms of H3K9 were increased in D2-containing cells. In addition to methylation at lysine residues, cocaine can also alter arginine methylation (Damez-Werno et al., 2016). Specifically, cocaine SA led to significant decreases in protein-R-methyltransferase-6 (PRMT6) and its associated histone mark, asymmetric dimethylation of R2 on histone H3 (H3R2me2a) in rodents (Damez-Werno et al., 2016). Humans who suffered from cocaine use disorder also showed similar changes in their NAc.
5. Involvement of DNA methylation and hydroxymethylation in models of addiction
Because DNA methylation plays important roles in cognitive functions such as learning and memory (Jobe and Zhao, 2017) that are impacted by drugs of abuse (Bisagno et al., 2016), it was highly likely that these drugs might influence the expression of DNMTs. An acute cocaine injection increased DNMT3A and DNMT3B mRNA levels in the NAc (Anier et al., 2010). LaPlant et al. (2010) reported that DNMT3A mRNA was up-regulated at 4 h but down-regulated at 24 h after a single cocaine injection. Cocaine also increased DNA hypermethylation and increased binding of methyl CpG binding protein 2 (MeCP2) at the protein phosphatase-1 catalytic subunit (PP1c) promoter associated with decreased PP1C mRNA levels (Anier et al., 2010). Repeated cocaine injections also increased DNMT1, DNMT3A, and DNMT3B mRNA levels in the NAc (Anier et al., 2018). Interestingly, mice that self-administered cocaine showed significant decreases in DNMT1 and DNMT3A mRNAs measure 24 h after the last session (LaPlant et al., 2010). Using rats that self-administered cocaine, Wright et al. (2015) found significant increases of DNMT3A in the NAc but not in the prefrontal cortex. Of related interest, rats that underwent cocaine SA also exhibited increased MeCP2 expression in the dorsal striatum (Im et al., 2010). Using genome-wide sequencing, Baker-Andresen et al. (2015) found that rats that self-administered cocaine exhibited 29 differentially methylated regions in a persistent manner in comparison to saline- or passive cocaine-treated animals, with five regions being demethylated and the rest hypermethylated. Methamphetamine also influences the expression of DNMT gene expression. For example, Numachi et al. (2007) repeated injections of the drug increased DNMT1 expression in the NAc and dorsal striatum of Fisher rats. In addition, Jayanthi et al. (2019) have found that methamphetamine SA is accompanied by increased DNA methylation at the DNA sequences of several genes that code for potassium channels.
Interestingly, DNA hydroxymethylation marks have also been investigated after administration of cocaine and methamphetamine. Global levels of 5-hmC were increased after repeated cocaine administration (Anier et al., 2018). Ploense et al. (2018) found that limited access to cocaine decreased DNA hydroxymethylation within the Homer2 promoter in rats (Sadakierska-Chudy et al., 2017b). also reported that abstinence from cocaine was associated with increased global 5-hydroxymethylcytosine (5-hmC) levels and increased Tet 3 transcript levels in the hippocampus. Cadet et al. (2017) used an animal model of drug SA wherein footshocks were used to suppress methamphetamine SA in some but not in other rats. They found that the animals that suppress their intake of the drug had much higher levels of DNA hydroxymethylation at the sequences of several reward-related genes including potassium channels in the NAc. Injections of methamphetamine also produced DNA hypomethylation at sites near the corticotrophin (CRH) transcription start site (TSS) and at intragenic vasopressin (AVP) sequences. methamphetamine also increased DNA hydroxymethylation at these sites. Interestingly, there was increased protein expression of ten-eleven-translocation (TET) enzymes that catalyze DNA hydroxymethylation. Importantly, the TET inhibitor, 1,5-isoquinolinediol (IQD), blocked methamphetamine-induced increases in Crh and Avp mRNA expression.
Changes in DNA methylation have also been reported after opioid exposure. For example, Kozlenkov et al. (2017) reported 1298 differentially methylated CpG sites (DMSs) between heroin users and controls. Hyper-DMSs were enriched in gene bodies and exons but depleted in promoters while hypo-DMSs were enriched in promoters and enhancers. In addition, hyper-DMGs showed preference for genes involved in axonogenesis- and synaptic functions. Similarly, post-mortem brain samples from long-term opioid abusers have been reported to exhibit reduced mu-opioid receptor (OPRM1) expression that was related to altered methylation at a CpG site in the functional SNP variant of the OPRM1 (118A > G) gene. These changes in methylation were also accompanied with abnormal compensatory mechanisms that would have occurred with chronic opioid exposure (Oertel et al., 2012). Besides DNA methylation, epigenetic regulation via repressive histone methylation has also been reported after repeated morphine exposure (Koo et al., 2015). Moreover, the authors reported that chronic morphine administration caused behavioral sensitization that was associated with increased binding of H3K27me3 at the Bdnf-p2 promoter and secondary decreased Bdnf expression in the ventral tegmentum (VTA) (Koo et al., 2015). Taken together, these results implicate drug-induced methylation as a regulator of gene expression and behavioral changes associated with drug taking behaviors.
6. Concluding remarks and future directions
The present literature on the effects of drugs of abuse on epigenetic markers supports the idea that these drugs can have both acute and persistent effects on the expression of several enzymes involved in causing post-translational histone modifications and DNA methylation/hydroxymethylation in the brains of rodents. These changes are accompanied by differential changes in the abundance of modified histones on the promoters or bodies of genes that have been implicated in animal models of addiction. These observations are interesting and are remarkable in terms of supporting the notion that long-term neuroadaptations are responsible for the long-lasting behavioral effects of drugs of abuse. Although these findings are very interesting, it remains to be determined to what extent, they will be translatable to human conditions. Studies being conducted in models that use footshocks to separate groups of rodents that use cocaine, methamphetamine, and opioids into resilient and susceptible rodents may enhance the ability of scientists to translate these results into the development of pharmacotherapeutics that might revert some of the epigenetic changes observed in the brain of drug exposed animals. These novel studies promise to provide a better window to the long-term impact of drug taking behaviors and of withdrawal/abstinence on epigenetic markers (see scheme in Fig. 2). There is, however, a very important need to expand these types of investigations to post-mortem tissues in a fashion similar to studies of post-mortem tissues of patients who suffer from Schizophrenia, major affective disorder, Alzheimer's and Parkinson's diseases, and normal and abnormal aging. Epigenetic studies of post-mortem tissues may help to further clarify what animal models of addiction are more relevant to human situations.
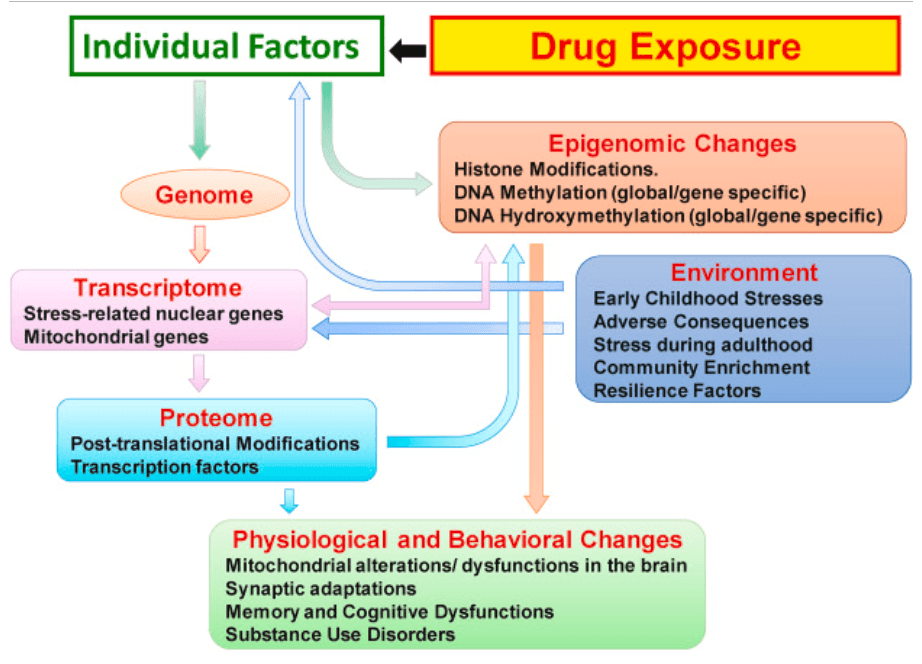
Fig. 2. The scheme described here indicates that individual licit and illicit drugs can impact individuals in very specific ways as reported in the clinical literature. Not all individuals who are exposed to drugs become addicted to these drugs and their responses to drugs depend on genetic and environment factors that either protect them or help to escalate their uses of specific initially rewarding agents. This figure also suggests the need to identify the differential and progressive epigenetic and transcriptional consequences of drug exposure that determine paths to compulsive drug use despite adverse consequences or sustained drug use without developing a diagnosis of substance use disorder. The schema also suggests that treatment approaches will depend on pharmacological agents that can block the further development of epigenetic changes or reverse already established epigenetic modifications.
It is also important to conclude with some suggestion about the potential use of drugs that might impact epigenetic mechanisms in the brain. This is important because these drugs will not impact epigenetic mechanisms and gene transcription in the central nervous system but also in various peripheral organs. At present, there are several drugs that being used as anticancer therapeutics (Nepali and Liou, 2021). These drugs can impact many of the targets that we have discussed in this review including histone acetylation and methylation as well as DNA methylation (Nepali and Liou, 2021; Sarno et al., 2021). Given the levels of plasticity associated with the epigenome, the use of similar compounds in therapeutic interventions against SUDs seems quite warranted. However, the translation of these ideas to clinical practice will require great investments of resources to investigate the precise doses of specific epigenetic drugs and their combinations to treat specific SUD. Moreover, it will be important to carefully calculate the benefits of using these drugs in contrast to adverse events encountered during their clinical use. Some of these negative consequences may be avoided by the use of pro-drugs that are transformed to their active forms in the brain. Because drugs of abuse appear to influence multiple epigenetic targets, it will be important to generate therapeutic mixtures that might influence different targets at the same time. This poly-pharmacological approach has been used successfully in medicine and psychiatry for many years. Finally, the future use of epigenetic agents promises to add in the clinicians’ armamentarium against SUDs because the repeated oral intake or intravenous injections of licit and illicit drugs are associated with abnormalities in multiple epigenetic pathways. This approach may be more productive beyond approaches that use single drugs that block or stimulate single neurotransmitter receptors.